By Eduardo Moralejo & Àlex Giménez-Romero
The effect of climate change and globalization is expected to have a considerable impact on agriculture, affecting both crop distribution and associated harmful organisms1. Understanding how these two processes act on the geographical expansion of pests and pathogens is not an easy task because both processes are indirectly related. Furthermore, since the early 1990s, both the international trade in plants and the increase in average temperature have increased exponentially, so it is not surprising that the underlying causes of changes in the distribution of pests and pathogens are often confused. Fortunately, powerful new molecular and phylogenetic tools allow us to trace the origin of the introduction of harmful organisms. However, we know anything about the failed introduction events, which raises doubts about whether the current distribution is due to climatic or ecological factors or simply because of the particular spatial distribution of introduction events.
We have asked ourselves this question to understand the current, past and future distribution of Pierce’s disease of grapevine, one of the most serious threats to world viticulture. To circumvent this dilemma, we have removed the effect of globalization from the system in the simplest way. We have assumed in the model that each year the pathogen is introduced and replenished in all parts of the planet, so that only two limiting parameters intervene in the simulations of the epidemiological model, the hourly temperature and the presence of insect vectors. In the article titled “Global predictions for the risk of establishment of Pierce’s disease of grapevine” published in Communications Biology we explain this approach2.

Figure: Pierce’s disease of grapevine ( (photo: Aaron Jacobson/UC Davis).
Pierce’s disease of grapevine is caused by the bacterium Xylella fastidiosa, a vascular pathogen of woody plants that can potentially infect more than 500 species3,4. The species is composed of three subspecies, fastidiosa, multiplex and pauca, within which there are more than 80 clonal lineages with diverse and usually different host ranges5. Pierce’s disease is caused by the sequence type ST1 and some close variants of the fastidiosa subspecies6. Until 2013 it was only known to occur in vineyards and almond plantations in North America. Today it is distributed in Taiwan7, possibly in Iran8, Israel9 and in Europe it has only been detected on the island of Mallorca in Spain10,11. Other X. fastidiosa subspecies have been found in Italy, France and Portugal since 2013, for which there is great concern from the European phytosanitary authorities4. Therefore, knowing the potential distribution of X. fastidiosa and the possible economic impact on European agriculture is a priority issue.
The pathogen X. fastidiosa was accidentally introduced to Mallorca through grafting material from infected almond trees brought from California, most likely around 199312. However, it was not until 2016 that it was identified as the causative agent of the massive death of almond trees in the island. The disease was confused with a problem caused by wood fungus and the effect of climate change13. The same clonal lineage of X. fastidiosa produces Pierce’s disease, however, no one realized that it had been in Mallorcan vineyards for many years, perhaps because the damage in general was not considerable.
The fact that X. fastidiosa is widely distributed in Mallorca has been used to carry out inoculations on more than 36 vine varieties in independent trials for three years under conditions similar to those in the field. The evolution of the symptoms of Pierce’s disease in each plant has been monitored and the hourly average temperature have been collected. These data have made it possible to model the development of symptoms of Pierce’s disease as a function of temperature accumulation during the growing season, adjusting the degree days to the cardinal growth temperatures of the bacteria in vitro14. In this way, a risk probability function for the development of Pierce’s disease has been obtained as a function of the accumulated modified growing degree-days (MGDD).
Figure: Xylella fastidiosa bacteria clogging a xylem vessel of a grape leaf.
On the other hand, the phenomenon of winter cold curing of infected vines in the USA is well known. Although the mechanism is not well understood, it is well established that temperature exposure limits the distribution of Pierce’s disease in relatively cold areas. In our work, the action of the cold has been modelled based on the accumulation of hours of cold below a base temperature of 6ºC in a similar way to the MGDD. We have searched globally for the relationship between the average minimum temperatures of the coldest month and the number of cold-degree days (CDD) units, conservatively assuming that the accumulation of CDD equivalent to -º1C of average minimum temperature eliminates ~75% of infections.
Transmission of X. fastidiosa in Pierce’s disease and other disease depends on the presence and abundance of insect vectors15. In the case of Europe, it seems that Philaenus spumarius is the only relevant vector of crop diseases caused by X. fastidiosa. In our work we have tried to indirectly estimate the basic reproductive number of the transmission of Pierce’s disease in Europe based on the ALSD epidemic outbreak in Mallorca. From the distribution and relative abundance maps of P. spumarius, it has been possible to infer how the vector populations modulate the maximum reproductive number. As we do not have exact data from other vectors, estimates from our model have been used to validate the distribution of Pierce’s disease in the US.
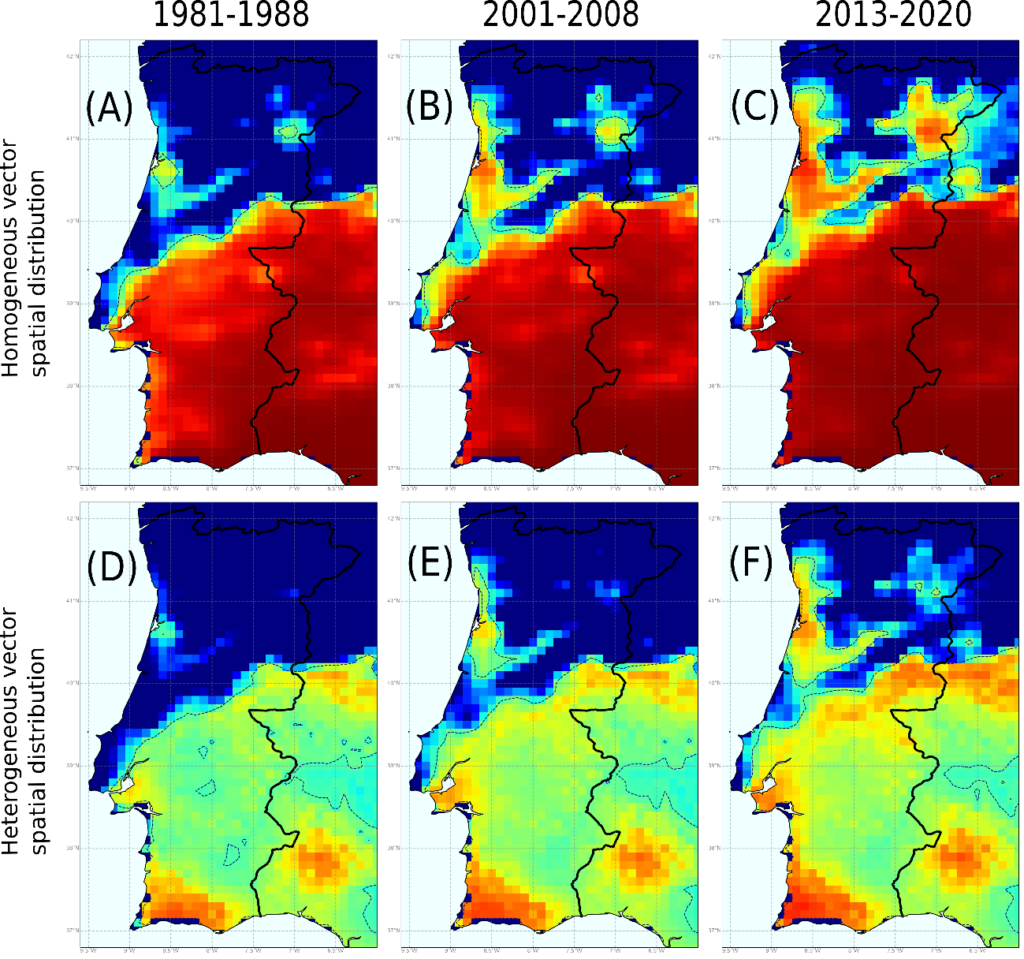
Figure. Risk of establishment of Pierce’s disease for the periods 1981-1988, 2001-2008 and 2013-2020. Panels (A)-(C) were simulated using a homogeneous spatial distribution of the vector, being R0 = 5 in all cells. Panels (E)-(F) were simulated with a heterogeneous spatial distribution of the vector, being R0_max = 5 where the density of the vector is maximal.
Our final model combines a climatic and transmission layers that determine the evolution of the disease over time, forcing the introduction of the pathogen each year. Based on annual hourly surface temperature data for the period 1980-2020 from Copernicus ERA5-Land, forward simulations were made with real data on the exponential growth or decrease of epidemic outbreaks. The model captures with very high reliability the places where Pierce’s disease exists in the US. In Europe, because vector information can be entered, the maps show more pronounced risk gradients, indicating that the greatest risk is in the Mediterranean islands and in Mediterranean continental areas where the pathogen has already been established. Our model has the virtue of giving quantifiable risks based on the exponential growth of infected vine plants, which shows the relative pressure of the disease where it is established.
The model is dynamic; therefore, it can be observed how the risk evolves over time with the increase in average temperature. In the last decade it is perceived that Atlantic mild-temperature wine-producing areas without risk or in a transition zone before 1990s such as Bordeaux or the Portuguese Duero riverbank, are evolving towards low-risk areas and that the future projections of the model towards 2050 indicate that the risk could reach moderate. It is in these areas where the possible introduction of the pathogen must be carefully monitored. It is in our hands to take measures to prevent the entry of plant material infected with X. fastidiosa that can be dispersed through local vectors and that can develop epidemics in vineyards in the future.
Read more at: https://rdcu.be/c6qk2

Eduardo Moralejo has a degree in Biology from the University of the Balearic Islands. He works part-time at TRAGSA as a contract researcher on the control of Xylella fastidiosa in the Balearic Islands. He has published more than 30 articles in international journals on the taxonomy, pathology and evolutionary ecology of the genus Phytophthora and the risk of introducing plant pathogens through international plant trade.

Àlex Giménez-Romero has a degree in Physics from the University of Barcelona and a master in Physics of Complex Systems from the University of the Balearic Islands. As a PhD student at IFISC, he studies ecosystems threatened by emergent epidemics and climate change through the lens of Complex Systems science. He has publications in international journals on modelling mollusc and plant epidemics and AI applied to ecology.
References
- Pautasso, M., Döring, T. F., Garbelotto, M., Pellis, L. & Jeger, M. J. Impacts of climate change on plant diseases—opinions and trends. European Journal of Plant Pathology 133, 295–313 (2012).
- Gimenez-Romero, A. et al. Global predictions for the risk of establishment of Pierce’s disease of grapevines. Communications Biology 1389 10.1038/s42003-022-04358-w (2022).
- Hopkins, D. L. & Purcell, A. H. Xylella fastidiosa: cause of Pierce’s disease of grapevine and other emergent diseases. Plant disease 86, 1056–1066 (2002).
- Authority (EFSA), E. F. S., Delbianco, A., Gibin, D., Pasinato, L. & Morelli, M. Update of the Xylella spp. host plant database–systematic literature search up to 30 June 2021. EFSA Journal 20, e07039 (2022).
- Vanhove, M. et al. Genomic diversity and recombination among Xylella fastidiosa subspecies. Applied and environmental microbiology 85, e02972-18 (2019).
- Nunney, L. et al. Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce’s disease of grapevine in the US. PLoS One 5, e15488 (2010).
- Leu, L. S. & Su, C. C. Isolation, cultivation, and pathogenicity of Xylella fastidiosa, the causal bacterium of pear leaf scorch disease in Taiwan. Plant Disease 77, 642–646 (1993).
- Amanifar, N., Taghavi, M., Izadpanah, K. & Babaei, G. Isolation and pathogenicity of Xylella fastidiosa from grapevine and almond in Iran. Phytopathologia Mediterranea 318–327 (2014).
- Zecharia, N. et al. Xylella fastidiosa outbreak in Israel: population genetics, host range and temporal and spatial distribution analysis. Phytopathology (2022).
- Gomila, M. et al. Draft genome resources of two strains of Xylella fastidiosa XYL1732/17 and XYL2055/17 isolated from Mallorca vineyards. Phytopathology 109, 222–224 (2019).
- Moralejo, E. et al. Insights into the epidemiology of Pierce’s disease in vineyards of Mallorca, Spain. Plant Pathology 68, 1458–1471 (2019).
- Moralejo, E. et al. Phylogenetic inference enables reconstruction of a long-overlooked outbreak of almond leaf scorch disease (Xylella fastidiosa) in Europe. Communications biology 3, 1–13 (2020).
- Olmo, D. et al. Landscape epidemiology of Xylella fastidiosa in the Balearic Islands. Agronomy 11, 473 (2021).
- Feil, H. & Purcell, A. H. Temperature-dependent growth and survival of Xylella fastidiosa in vitro and in potted grapevines. Plant Disease 85, 1230–1234 (2001).
- Cornara, D. et al. Transmission of Xylella fastidiosa by naturally infected Philaenus spumarius (Hemiptera, Aphrophoridae) to different host plants. Journal of Applied Entomology 141, 80–87 (2017).

